Isostable DNA Duplexes: Difference between revisions
(Created page with " '''Isostable DNA Duplexes''' The stability of DNA duplexes depends strongly on the sequence. Because G:C base pairs are considerably more stable than A:T base pairs, the G:C content determines how high a temperature is required for dissociation of the strands forming a duplex. The higher the G:C content, the greater the thermal stability. The sequence dependence of the stability makes it difficult to detect A/T-rich sequences in a genomic context, e.g. in diagnostic...") |
(Removed the pi-pi stacking interaction between adenine and 6-ethynylpyridone.) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Isostable DNA Duplexes == | |||
[[File:Hypoxanthine Cytosine base pair new.png|right|thumb|Non-canonical base pair between hypoxanthine and cytosine.]] | |||
[[File:6-Ethynylpyridone Adenine base pair.png|right|thumb|Non-canonical base pair between adenine and 6-ethynylpyridone.]] | |||
The stability of DNA duplexes depends strongly on the sequence. Because G:C base pairs are considerably more stable than A:T base pairs, the G:C content determines how high a temperature is required for dissociation of the strands forming a duplex. The higher the G:C content, the greater the thermal stability. The sequence dependence of the stability makes it difficult to detect A/T-rich sequences in a genomic context, e.g. in diagnostic or analytical tests. To overcome this problem, the concept of 'isostable DNA' was developed. In isostable DNA, the thermal stability of duplexes is independent of the G/C content. One way to accomplish this is to use non-canonical nucleobases. For example, guanine may be replaced by hypoxanthine to weaken the base pair with C, or thymine may be replaced by 6-ethynylpyridone as nucleobase surrogate to get a more stable base pair. | |||
== References == | |||
[1] H.K. Nguyen, O. Fournier, U. Asseline, D. Dupret, N.T: Thuong, Smoothing of the thermal stability of DNA duplexes by using modified nucleosides and chaotropic agents. ''Nucleic Acids Res.'' '''1999''', ''27'', 1492-1498. https://doi.org/10.1093%2Fnar%2F27.6.1492 | |||
[2] C. Ahlborn, K. Siegmund, C. Richert, Isostable DNA. ''J. Am. Chem. Soc.'' '''2007''', ''129'', 15218-15232. https://doi.org/10.1021/ja074209p | |||
[3] M. Minuth, C. Richert, A nucleobase analogue that pairs strongly with adenine. ''Angew. Chem. Int. Ed.'', '''2013''', ''52'', 10874-10877. https://doi.org/10.1002/anie.201305555 | |||
Latest revision as of 08:36, 6 September 2024
Isostable DNA Duplexes
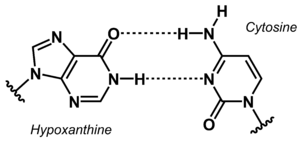
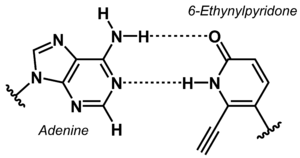
The stability of DNA duplexes depends strongly on the sequence. Because G:C base pairs are considerably more stable than A:T base pairs, the G:C content determines how high a temperature is required for dissociation of the strands forming a duplex. The higher the G:C content, the greater the thermal stability. The sequence dependence of the stability makes it difficult to detect A/T-rich sequences in a genomic context, e.g. in diagnostic or analytical tests. To overcome this problem, the concept of 'isostable DNA' was developed. In isostable DNA, the thermal stability of duplexes is independent of the G/C content. One way to accomplish this is to use non-canonical nucleobases. For example, guanine may be replaced by hypoxanthine to weaken the base pair with C, or thymine may be replaced by 6-ethynylpyridone as nucleobase surrogate to get a more stable base pair.
References
[1] H.K. Nguyen, O. Fournier, U. Asseline, D. Dupret, N.T: Thuong, Smoothing of the thermal stability of DNA duplexes by using modified nucleosides and chaotropic agents. Nucleic Acids Res. 1999, 27, 1492-1498. https://doi.org/10.1093%2Fnar%2F27.6.1492
[2] C. Ahlborn, K. Siegmund, C. Richert, Isostable DNA. J. Am. Chem. Soc. 2007, 129, 15218-15232. https://doi.org/10.1021/ja074209p
[3] M. Minuth, C. Richert, A nucleobase analogue that pairs strongly with adenine. Angew. Chem. Int. Ed., 2013, 52, 10874-10877. https://doi.org/10.1002/anie.201305555